Deep Learning Allows for Digital Labeling of Multiple Cellular Structures
BY: GINA MANTICA
Stem cells can help repair damaged tissues, but only if they aren’t damaged, themselves. Some techniques to visualize the intricate structures of stem cells are time-consuming and invasive, while others require the cells to be killed. But machine learning can significantly cut down the time it takes to visualize the different parts of cells, while keeping the cells alive and intact.
With support from the Hariri Institute, Research Fellow Lei Tian turned his idea for an informative, label-free cellular imaging technique into a reality. Tian and colleagues used a combination of reflectance light microscopy and machine learning to generate images of single cells that accurately depict their tiny, cellular components. Their findings were recently published in Science Advances.

Tian and colleagues’ method allows for a more holistic picture of a single cell without the use of any harsh chemicals or gene editing. By combining new microscopy techniques with machine learning, scientists can quickly and efficiently examine hundreds of individual cells at an incredibly fine level of detail.
The researchers used reflectance light microscopy to look at the microscopic details of a cell’s intricate structures. Reflected light bounces off the top of specimens, while transmitted light illuminates specimens from below. Though both methods can be used to look at individual cells, reflectance light microscopy helps researchers detect structural changes on the “top”, or surface, of a cell that can be hard to see with transmitted light.
To capture a single cell’s elaborate interior, Tian and colleagues used multiple light-emitting diodes (LEDs) in their microscope. The LEDs were arranged in a grid so that the researchers could change the number and direction of lights used to illuminate the cells. Changing the brightness and angle of the LEDs affects how the light reflects off the surface of a cell and in turn, how the cell looks through the microscope. Researchers can better capture the rich details of a cell’s microscopic components, like its nucleus or membrane, by viewing the cell from different perspectives. And the more detailed the images are, the better the deep learning algorithm can be at “digitally labeling” tiny structures within an individual cell.
The researchers used their microscope to look for four different cellular structures–deoxyribonucleic acid (DNA), Golgi apparatus, endosomes, and actin. The team also looked for signs that a cell could be replicating or dying. They then trained their deep learning model on images of cells stained with fluorescent markers for each of these six features.
After training, the team tested their deep learning framework on images of cells without the fluorescent markers. They found that their model can accurately predict and label where the four different structures should be in individual cells. And, their model can combine multiple predictions into a single image. So rather than having separate images of a cell’s DNA, Golgi apparatus, endosomes, or actin, the team can look at all these different cellular components together.
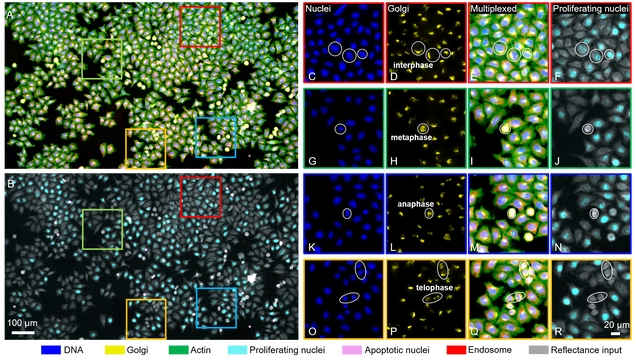
This “multiplexing”, or combining of different labels into a single image, is so accurate that scientists can even use the labeled images to determine a cell’s phase in the cell cycle. Tian and colleagues’ deep learning model can detect slight changes to the Golgi apparatus that happen when a cell transitions into mitosis, or cellular division.
“Multiplexing can be hard. This technique could be useful in high-throughput applications to digitally fuse many different fluorescent labels together and give you a holistic picture of large populations of cells,” said Tian, “The Hariri Institute’s award gave our team a nice boost to get this project off the ground.”
Interested in learning more about the transformational science happening at the Hariri Institute? Sign up for our newsletter here.
