News

BME Researchers Top Field in Predicting Protein-Protein Interactions
“Protein-protein interactions help you to figure out the mechanisms of life, and provide important drug targets." More

Promotions for ENG Faculty
"They exemplify each day the depth and excellence of Boston University’s talented academic community.” More

An Art of Iteration
After a career designing mammogram machines and pacemakers, she is making experimental works of art meant to convey universal emotions. More

Developing a Life-Saving Option for a Rare Childhood Disorder
As a Hartwell Investigator, Samagya Banskota and her collaborators will leverage a cutting-edge genome editing technique. More

Spotting Signs of Cancer When There’s Still Time
With a bold plan for a new imaging platform, Liangliang Hao has earned a 2025 Beckman Young Investigator Award. More

Novel Solutions to Health Disparities
A skin tone colorimeter and a cornea transplant training tool took first prizes in the Design-A-Thon. More

Disentangling Behavior: Cognition and Movement
Are cognitive processes, such as planning an errand or trying to recall a name, separable from related muscle movements? More

The Spring of Hope
The Class of 2025 celebrates Commencement. More

Thomas A. Edison Patent Award for Xin Zhang
Zhang is being recognized for her pioneering contributions to the field of metamaterials. More
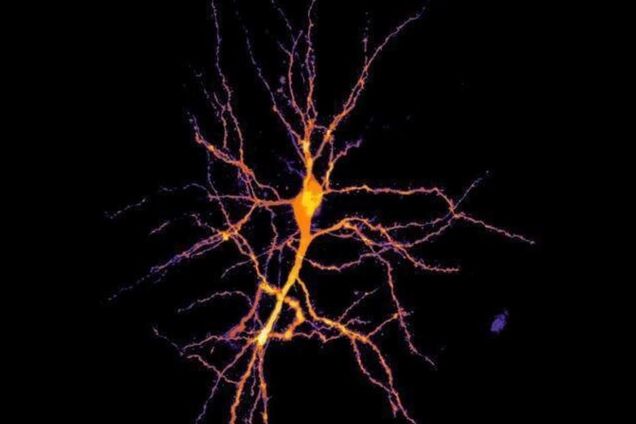
Combining Cutting-Edge Techniques To Study How Brain Cells Function Together
"Imagine a therapy for patients with memory deficits, in which their brain is stimulated in a precisely timed manner to improve their recall ability." More
