NF-kappaB Transcription Factors
Dr. Thomas D. Gilmore
Biology Department, Boston University
5 Cummington Mall, Boston, Massachusetts 02215-2406, USA
(617) 353-5444 or 5445; fax (617) 353-6340
The Rel/NF-kappaB Signal Transduction Pathway
Rel or NF-kappaB (NF-kB) proteins comprise a family of structurally-related eukaryotic transcription factors that are involved in the control of a large number of normal cellular and organismal processes, such as immune and inflammatory responses, developmental processes, cellular growth, and apoptosis. In addition, these transcription factors are persistently active in a number of disease states, including cancer, arthritis, chronic inflammation, asthma, neurodegenerative diseases, and heart disease (see DISEASES link).
Rel/NF-kB transcription factors include a collection of proteins, with functions conserved from some single-celled organisms, sponges, sea anemones, and the fruit fly Drosophila melanogaster to humans. That is, NF-kB homologs have been found in organisms as simple as Cnidarians (e.g., sea anemones, corals, jellyfish, hydra), Porifera (sponges) and the single-celled eukaryotes Capsaspora owczarzaki and choanoflagellates Among the commonly studied model organisms, these transcription factors are notably absent in yeast and the nematode Caenorhabditis elegans (the latter having apparently lost the pathway during evolution); in part, the absence of the NF-kB pathway in yeast may be because one of the primary roles of these factors is to control a variety of physiological aspects of immune and inflammatory responses.
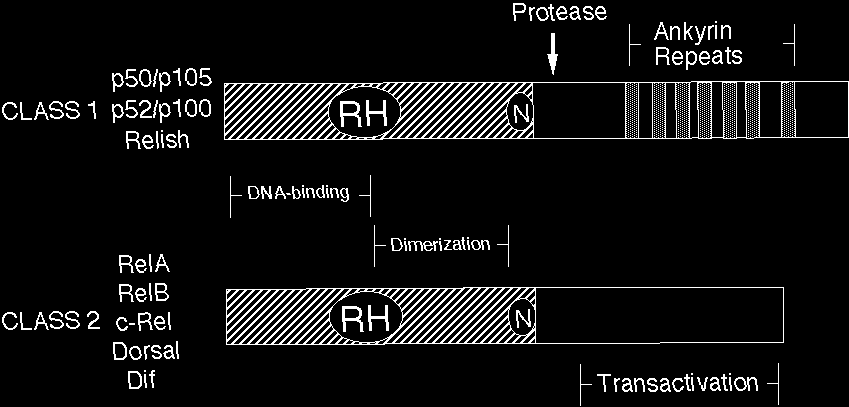
Rel/NF-kB proteins are related through a highly conserved DNA-binding/dimerization domain called the Rel homology (RH) domain. However, Rel/NF-kB proteins can be divided into two classes based on sequences C-terminal to the RH domain (Figure 1). Members of one class (the NF-kB proteins p105, p100, and Drosophila Relish) have long C-terminal domains that contain multiple copies of ankyrin repeats, which act to inhibit these molecules. Members of the NF-kB class become active, shorter DNA-binding proteins (p105 to p50, p100 to p52) by either limited proteolysis or arrested translation. As such, members of this first class are generally not activators of transcription, except when they form dimers with members of the second class of Rel/NF-kB transcription factors. The second class (the Rel proteins) includes c-Rel (and its retroviral homologue v-Rel), RelB, RelA (p65), and the Drosophila Dorsal and Dif proteins. This second class of Rel proteins contains C-terminal transcription activation domains, which are often not conserved at the sequence level across species, even though they can activate transcription in a variety of species. The cDNA and predicted protein sequences of Rel/NF-kB transcription factors can be rapidly accessed via this site (see SEQUENCES link).
Rel/NF-kB transcription factors bind to 9-10 base pair DNA sites (called kB sites) as dimers. All vertebrate Rel proteins can form homodimers or heterodimers, except for RelB, which can only form heterodimers. This combinatorial diversity contributes to the regulation of distinct, but overlapping, sets of genes, in that the individual dimers have distinct DNA-binding site specificities for a collection of related kB sites. The term NF-kappaB commonly refers specifically to a p50-RelA heterodimer, which is one of the most avidly forming dimers and is the major Rel/NF-kB complex in most cells. The x-ray crystallographic structures of several Rel/NF-kB dimers on DNA (including p50-p50, p65-p65, p50-p65, c-Rel-c-Rel, p50-p65-IkB) have now been solved, and these structures can be accessed from this site (see STRUCTURES link)
The activity of NF-kB is primarily regulated by interaction with inhibitory IkB proteins. As with the Rel/NF-kB proteins, there are several IkB proteins, which have different affinities for individual Rel/NF-kB complexes, are regulated slightly differently, and are expressed in a tissue-specific manner. The IkB proteins include, at least, p105, p100, IkBa, IkBb, IkBg, IkBe, IkBz, Bcl-3, and the Drosophila Cactus protein. The cDNA and predicted protein sequences of these IkBs can be obtained through this site (see SEQUENCES link).
The best-studied NF-kB-IkB interaction is that of IkBa with the NF-kB p50-RelA dimer. This interaction blocks the ability of NF-kB to bind to DNA and results in the NF-kB complex being primarily in the cytoplasm due to a strong nuclear export signal in IkBa. That is, the NF-kB-IkBa complex is continuously shuttling between the nucleus and the cytoplasm, but its rate of nuclear export exceeds its rate of import and thus the complex is generally cytoplasmic. From biochemical studies and direct structural determinations (see links at this site), it is clear that IkBa makes multiple contacts with NF-kB. These interactions cover sequences of NF-kB that are important for DNA binding. In contrast, when IkBb interacts with the NF-kB complex, the complex is retained in the cytoplasm (i.e., does not undergo nucleo-cytoplasmic shuttling). Thus, not all NF-kB-IkB interactions are the same.
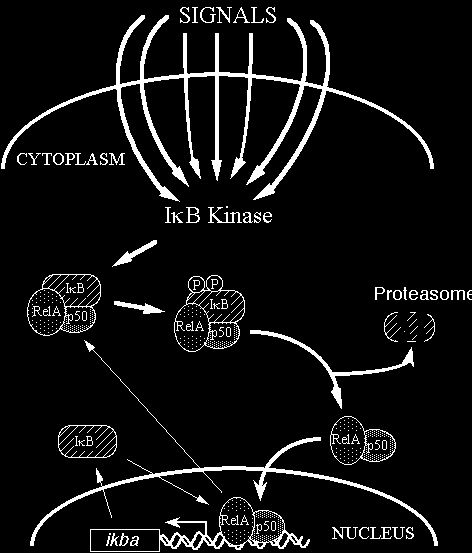
In most cells, NF-kB is present as a latent, inactive, IkB-bound complex in the cytoplasm. When a cell receives any of a multitude of extracellular signals (see INDUCERS link), NF-kB rapidly enters the nucleus and activates gene expression (see TARGET GENES link). Therefore, a key step for controlling NF-kB activity is the regulation of the IkB-NF-kB interaction. Many of the molecular details of this control are now understood (Figure 2). Almost all signals that lead to activation of NF-kB converge on the activation of a high molecular weight complex that contains a serine-specific IkB kinase (IKK). IKK is an unusual kinase in that in most cells IKK contains (at least) three distinct subunits: IKKalpha, IKKbeta and IKKgamma. IKKa and IKKb are related catalytic kinase subunits, and IKKg (aka NEMO) is a regulatory subunit that serves as a sensing scaffold and integrator of upstream signals for activation of the catalytic subunits. In the classical or canonical pathway, activation of IKK complex leads to the phosphorylation by IKKb of two specific serines near the N terminus of IkBa, which targets IkBa for ubiquitination (generally by a complex called beta-TrCP) and degradation by the 26S proteasome. In the non-canonical (or alternative) pathway, the p100-RelB complex is activated by phosphorylation of the C-terminal region of p100 by an IKKa homodimer (lacking IKKgamma), which leads to ubiquitination followed by degradation of the p100 IkB-like C-terminal sequences to generate p52-RelB. In either pathway, the unmasked NF-kB complex can then enter the nucleus to activate target gene expression. In the classical pathway, one of the target genes activated by NF-kB is that which encodes IkBa. Newly-synthesized IkBa can enter the nucleus, remove NF-kB from DNA, and export the complex back to the cytoplasm to restore the original latent state. Thus, the activation of the NF-kB pathway is generally a transient process, lasting from 30-60 minutes in most cells.
A variety of recent evidence, however, indicates that the control of the NF-kB pathway is more complex than simply IKK-mediated regulation of the IkB-NF-kB interaction. For example, RelA and p50 are regulated by ubiquitination, acetylation, methylation, phosphorylation, oxidation/reduction, and prolyl isomerization. Moreover, as a consequence of induction of NF-kB activity (at least by tumor necrosis factor) IKKa is also induced to enter the nucleus where it becomes associated with kB site promoters/enhancers to phosphorylate histone H3 which enhances the transcription of kB site-dependent genes. Finally, proteins in the NF-kB signaling pathway participate in a number of protein-protein interactions with non-NF-kB proteins (see PROTEIN-PROTEIN INTERACTIONS link).
In some normal cells, such as B cells, some T cells, Sertoli cells and some neurons, NF-kB is constitutively located in the nucleus. In addition, in many cancer cells (including breast cancer, colon cancer, prostate cancer, lymphoid cancers, and probably many others; see DISEASES link) NF-kB is constitutively active and located in the nucleus. In some cancers, this is due to chronic stimulation of the IKK pathway, while in other cases (such as some Hodgkin’s and diffuse large B-cell lymphoma cells) the gene encoding IkB can be mutated and defective. Moreover, several human lymphoid cancer cells have mutations or amplifications of genes encoding Rel/NF-kB transcription factors (esp REL in human B-cell lymhoma) and many multiple myelomas have mutations in genes encoding NF-kB signaling regulatory proteins that lead to constitutive activation of NF-kB. It is thought that continuous nuclear Rel/NF-kB activity protects cancer cells from apoptosis and in some cases stimulates their growth. Therefore, many current anti-tumor therapies seek to block NF-kB activity as a means to inhibit tumor growth or to sensitize the tumor cells to more conventional therapies, such as chemotherapy.
The Rel/NF-kB family is arguably the most-studied collection of eukaryotic transcription factors. For a collection of reviews on these transcription factors, the reader is directed to the November 22, 1999 and October 30, 2006 issues of Oncogene, which contain a series of reviews on Rel/NF-kB.
Our extensive knowledge of Rel/NF-kB signaling exposes also the reaches of our ignorance. We still have very little understanding of the complex in vivo dynamics of this pathway. For example, in most cell types and signaling conditions, it is still not known what are the contributions of specific Rel/NF-kB complexes (p50-RelA vs. p52-c-Rel vs. c-Rel-c-Rel) to most physiological responses. Over-expression studies in tissue culture almost certainly do not accurately reflect physiological signaling events. Similarly, what controls the balance between the levels of the various heterodimeric complexes in vivo is not known. Studies in Drosophila have elegantly shown that very small differences in nuclear concentrations of these factors, in their affinities for target DNA sites, and in cooperation or competition between Rel proteins and other transcription factors can have profound physiological consequences in organisms. Lastly, in many situations, it is not known how or which of the many genes induced by Rel/NF-kB factors in a given response contribute to that response. The development of methods to analyze genome-wide changes in gene expression (e.g., cDNA microarrays), which has already begun to uncover additional Rel/NF-kB-responsive genes, has helped to clarify which Rel/NF-kB target genes are activated in a given response.
As described above, the structures of several Rel/NF-kB dimers on DNA or bound to IkB are known. In all cases, these structures have been derived from molecules that contain almost exclusively residues from the RH domain. As such, these studies provide rather static glimpses of these factors at work. Several molecular and biochemical studies indicate that Rel dimers assume distinct conformations when bound to DNA versus as free or IkB-bound dimers or when bound to different kB sites. Moreover, such studies have also indicated that C-terminal residues influence sequences within the RH domain. Furthermore, there is surprisingly little information about how any of the Rel/NF-kB complexes actually activate transcription when bound to DNA: that is, what are the co-activators or basal factors with which they interact to activate transcription? Therefore, we cannot accurately simulate the dynamic nature of the complex as it releases from IkB, enters the nucleus, binds to DNA, and enhances gene expression; however,mathematical and computational modeling of the NF-kB pathway is beginning to address the dynamics of the pathway in response to various signals. In addition, recent studies suggest that NF-kB complexes bind to specific promoters in a dynamic on-off manner, with occupancy of a specific promoter sequence by an individual NF-kB dimer lasting on the order of seconds.
Although the discovery and characterization of the IkB kinase complex was a monumental step in our understanding of the regulation of this pathway, it raised almost as many questions as it has answered. For example, the following issues remain murky: 1) precisely which proteins are in the IKK complex in all cell types; 2) the exact size of the complex in all cell types; 3) what is the physiological relevance of phosphorylation by the IKK complex of substrates other than IkB; 4) how the various NF-kB and non-NF-kB activation pathways converge on IKK (for example, what and how many upstream kinases can activate IKK); 5) how is IKK activated by what appears to be induced clustering; 6) how is it that one subunit of this complex (IKKa) controls a specific developmental process, namely keratinocyte differentiation; 7) all of the other signaling pathways that crosstalk via or to IKK; 8) how do the two catalytic kinases within the IKK complex act on substrate proteins; and 9) how do reactive oxygen species and reactive Cysteine residues impact IKK activity. Recent X-ray crystal structural information on the IKK protein and components of the IKK complex may help answer some of these questions.
The study of v-Rel has unequivocally demonstrated that Rel/NF-kB transcription factors can be oncogenic, and one would like to know how the activating mutations in v-Rel have altered its structure as compared to c-Rel. However, v-Rel has accumulated so many activating mutations that it may not be a precise model for the role of these transcription factors in human cancers, where a single mutation (or gene amplification event) has occurred. Thus, in some cases, it is not known whether the rearrangements, mutations, and amplifications in Rel/NF-kB/IkB genes that have been repeatedly identified in several human cancers and the constitutive NF-kB signaling seen in certain human cancers or induced by oncogenic human viruses (e.g., EBV and HTLV-1) contribute to proliferation, abrogate growth suppression, influence the control of apoptosis, or affect all of these processes.
The extensive involvement of Rel/NF-kB transcription factors in human inflammation and disease establishes them as targets for therapeutics. Indeed, many common synthetic (e.g., aspirin), and traditional (e.g., green tea, curcumin) remedies target, at least in part, the Rel/NF-kB signaling pathway. However, there are over 800 compounds that have been shown to inhibit NF-kB signaling (see INHIBITORS at this site), and thus, the physiological or pharmacological utility of using any single compound for inhibition of NF-kB activity is a bit muddled. Nevertheless, our knowledge of the molecular details of this pathway is enabling the development of more specific and potent inhibitors of NF-kB signaling, and indeed, some NF-kB signaling inhibitors are entering clinical trials.
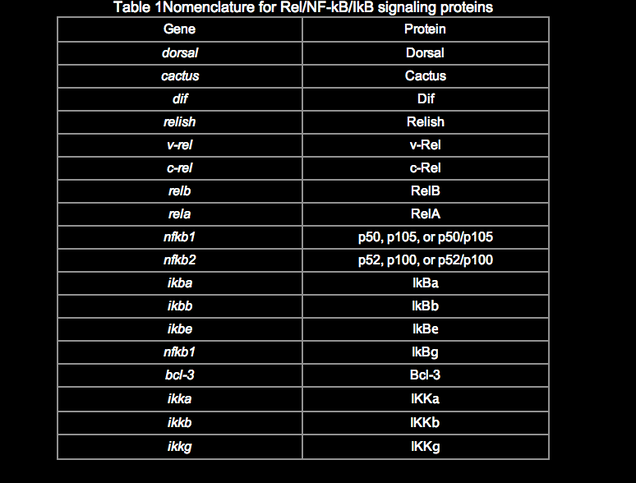
Among the many publications on this topic, there are inconsistencies in the naming of genes and proteins in the Rel/NF-kB pathway. Although a system of nomenclature for the Rel/NF-kB transcription factors and IkB proteins was established previously (Nabel and Verma, 1993), we use and suggest a slightly modified nomenclature (Table 1). The revised nomenclature reflects the new members of this pathway, common usage over the past several years, and at times my own judgment. In most cases, the choice was quite simple, although the p65 vs. RelA decision continues to be a thorny one.
Research in the Gilmore laboratory has been supported by the National Cancer Institute of the National Institutes of Health, the National Science Foundation, the American Cancer Society, the Cure for Lymphoma Foundation, the Leukemia Research Foundation, the Council for Tobacco Research, and Boston University. For more information on the Gilmore lab, go to the link for THE LAB.
Beyaert R (editor) (2004). Nuclear Factor-kappaB: Regulation and Role in Disease. Kluwer Academic Publishes, Dordrecht, The Netherlands. 426 pages
Gilmore TD (editor) (2006). NF-kB: from basic research to human disease. Oncogene (Reviews) 51: 6679-6899
Nabel GJ and Verma IM.(1993). Genes & Development 7: 2063
Perkins ND (2007) Integrating cell-signaling pathways with NF-kB and IKK function. Nature Reviews Molecular Cell Biology 8: 40-62
Figure 1 Structures of Rel family transcription factors. Shown are the generalized structures of the two classes of Rel transcription factors. All have a conserved DNA-binding/dimerization domain called the Rel homology (RH) domain, which also has sequences important for nuclear localization (N) and IkB inhibitor binding. Class I proteins have additional inserted sequences in the RH domain. The C-terminal halves of the class I Rel proteins have ankyrin repeat-containing inhibitory domains, which can be removed by proteasome-mediated proteolysis (PROTEASE). The C-terminal halves of the class II Rel proteins have transcriptional activation domains.
Figure 2 Rel/NF-kB signal transduction. In the classical pathway, various signals converge on activation of the IkB kinase (IKK) complex. IKK then phosphorylates IkB at 2 N-terminal serines, which signals it for ubiquitination and proteolysis. Freed NF-kB (p50-RelA, in this case) enters the nucleus and activates gene expression. One NF-kB target gene encodes IkB. The newly synthesized IkB can enter the nucleus, pull NF-kB off DNA, and export NF-kB back to its resting state in the cytoplasm. Thick lines indicate the activating pathway; thin lines indicate the inactivating pathway.