NSF Funds Investigation of Protein Structure and Dynamics
One of the great challenges in biological chemistry is understanding how protein structure informs function. Experimental studies have shown that by encapsulating proteins in surfactant micelles (nanoscale molecular aggregates, such as a droplet in a colloidal system) critical insights can be gained into protein structure, dynamics, and function. Currently, however, no one knows how the micelle environment influences the structure and dynamics of the encapsulated proteins.

The Chemical Theory, Models and Computational Methods Program of NSF’s Chemistry Division has funded Professor John Straub and his group to undertake a computational study to develop a fundamental understanding of the nature and potential applications of protein encapsulation in micelles.
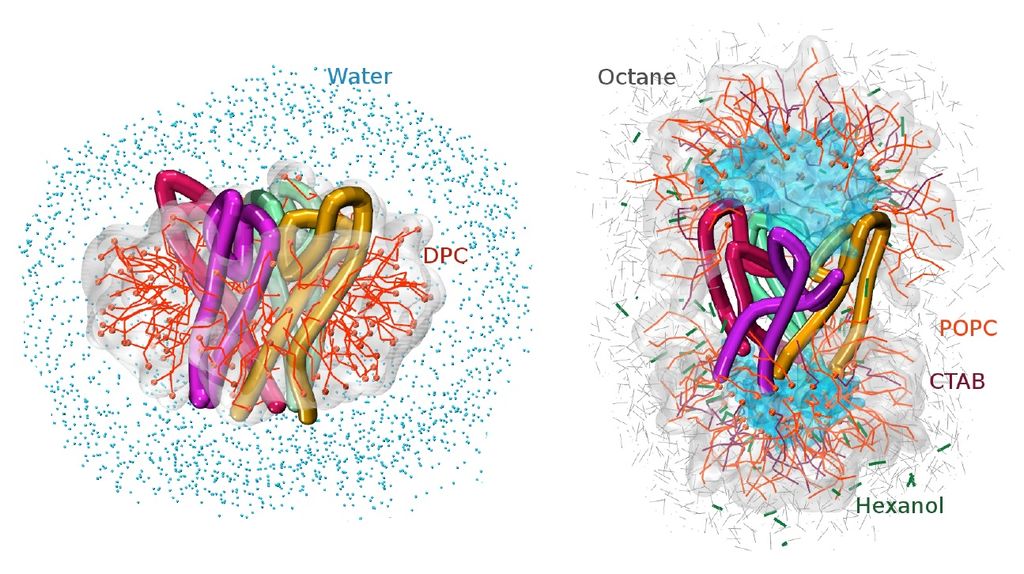
The project involves a systematic computational study of the effect of varying surfactant properties on the structure and dynamics of micelle-encapsulated proteins. Using a novel multiscale computational approach, the researchers will explore the influence of the choice of surfactant (anionic, cationic, and mixtures) on properties such as association equilibria of micelle-encapsulated proteins and internal solvation dynamics. They will directly compare their theoretical predictions with observables from NMR and vibrational spectroscopies. Their goal is to use the insights derived from these studies to propose improved micelle surfactants for future experimental use.
The broader impact of the study is the promotion of graduate education in computational biophysics through the development of a summer school for graduate training in state-of-the-art computational and experimental methods in the field of biomolecular structure and dynamics, to be hosted by the Telluride Science Research Center in Telluride, Colorado.