Research
The Mühlberger lab has a strong research interest in studying highly pathogenic viruses, including the filoviruses Ebola and Marburg virus. Work in the Mühlberger lab ranges from molecular biology studies on filoviral transcription and replication to dissecting host responses to filovirus infection. As virologists with an interest in emerging viruses, we are eager to collaborate with investigators who have expertise in different research areas, including tissue engineering, immunology, molecular imaging, and small RNA regulation to come up with innovative research approaches. The overarching goal of our work is to identify the determinants of virulence and pathogenicity in deadly viral diseases. Understanding these mechanisms paves the way towards the development of antiviral countermeasures.
Filovirus genome replication and transcription
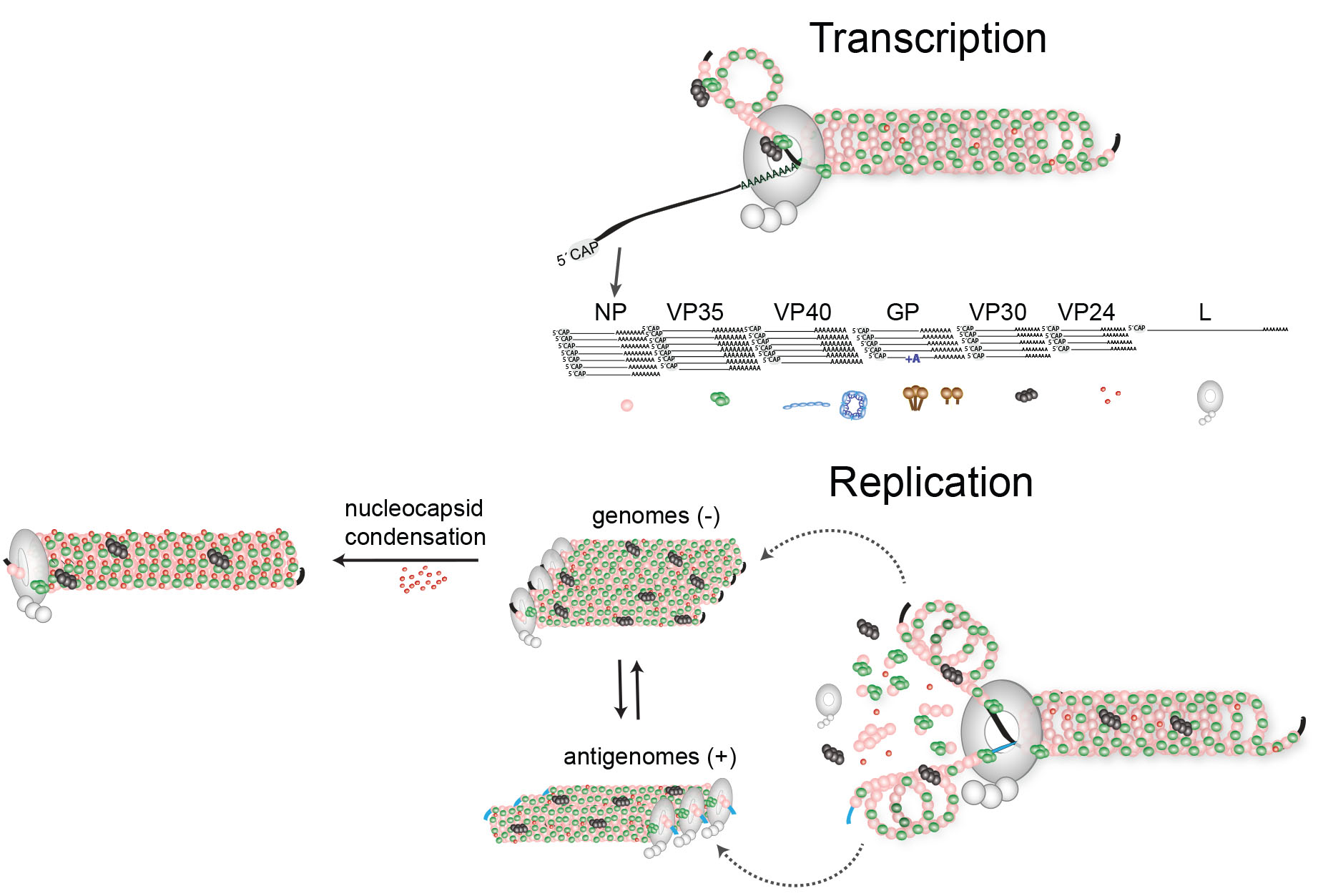
We have a couple of projects in the lab dealing with filovirus replication and transcription mechanisms. Jointly with Dr. Rachel Fearns, BU we aim to dissect the different steps of Marburg virus gene expression. How does the viral polymerase initiates transcription? What is the fate of the viral mRNAs in the infected cell? What are the temporal and spatial differences of viral transcription and replication? These are some of the questions we are tackling in this project.
High-throughput sequencing showed that filovirus genomes encode small RNAs. In collaboration with Dr. Daniel Cifuentes, BU we investigate the origin and the biologic function of these small virus-encoded RNAs in Ebola virus infection.
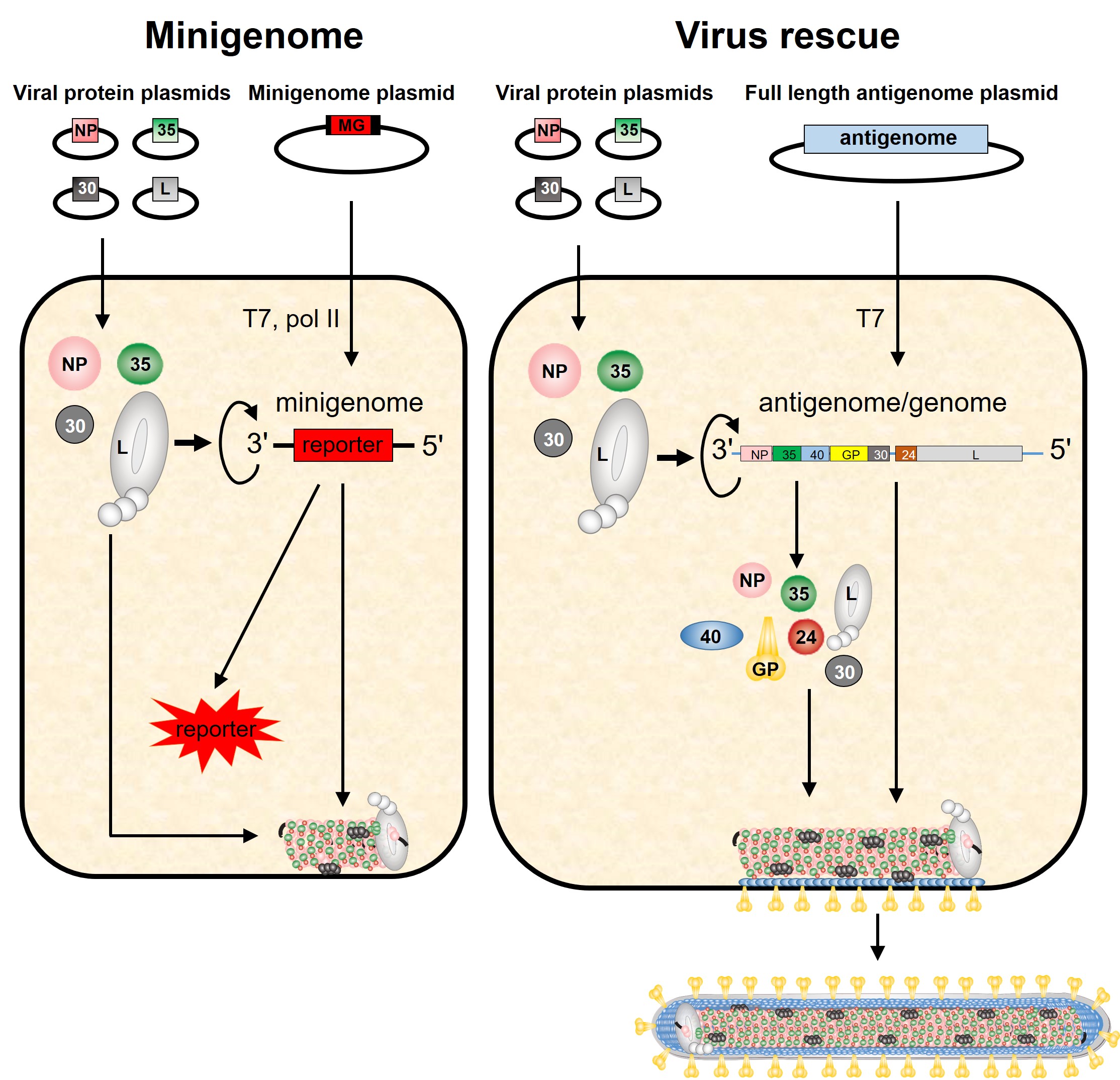
Research tools: Minigenome systems and recombinant viruses
The filovirus genome consists of a single RNA strand in negative orientation and is tightly associated with 5 viral proteins, the nucleocapsid proteins NP, VP35, VP30, L, and VP24. Four of these proteins (NP, VP35, VP30, L) are required for transcription and replication of the viral genome. The nucleocapsid proteins play a dual role in the viral replication cycle: they are structural components of the nucleocapsid complex and, therefore, involved in virus morphogenesis, and they catalyze replication and transcription of the RNA genome. Filoviruses are BSL-4 pathogens which makes it difficult to study viral replication mechanisms. To be able to do some mechanistic work a BSL-2, we have established minigenome systems for Ebola virus, Reston virus, Marburg virus, and Lloviu virus. We use the minigenome systems to study cis-acting signals on the RNA genome, including replication and transcription promoters, intergenic regions, transcription start- and stop signals, and to analyze the role of the viral proteins in replication and transcription.
In the BSL-4 lab, we can generate recombinant filoviruses using rDNA technology. For example, we can insert a reporter gene encoding a fluorescent protein in the filovirus genome. Cells infected with such a virus fluoresce. Recombinant viruses are useful tools to monitor viral spread.
Filoviruses, an expanding family
With the advent of high-throughput sequencing technology, the filovirus family has expanded. Newly discovered filoviruses include Bombali virus (BOMV), a new member of the Ebolavirus genus, Lloviu virus (LLOV), Měnglà virus (MLAV), and some fascinating fish filoviruses whose genome organization is very different from the “conventional” filovirus genomes. LLOV, MLAV, and the fish filoviruses are genetically distinct from ebola- and marburgviruses and have been assigned to distinct filovirus genera. While most of the known marburg- and ebolaviruses cause disease in humans, the pathogenic potential of the newly discovered filoviruses is not known.
The unexpected geographic and genetic diversity of filoviruses raises concerns about unpredictable outbreaks caused by the new members of the family. Having a thorough understanding of the filoviral replication mechanisms, including identifying similarities and differences in the replication strategies of different filoviruses, will be instrumental for designing and developing countermeasures in order to be prepared for potential outbreak scenarios. Of note, none of the new filoviruses were isolated from either an animal host or human patients – the available sequence information is based on viral RNA extracted from bats or fish and is incomplete with various amounts of terminal genomic sequences missing. This significantly hampers research on these viruses. To overcome this issue, we have established a minigenome system for LLOV that can be used to study LLOV transcription and replication. The data obtained with the mingenome system are instrumental for the rescue of infectious LLOV clones for pathogenesis studies.
Host responses to filovirus infection
Macrophages. A hallmark of fatal Ebola virus disease is a hyperactive inflammatory response, including a strong induction of interferon signaling pathways. Macrophages seem to play a pivotal role in the induction of this inflammatory response. In contrast to most cell types, macrophages are strongly activated when infected with Ebola virus and produce proinflammatory cytokines and chemokines. This proinflammatory response is triggered by the Ebola virus glycoprotein GP through the activation of Toll-like receptor 4 (TLR4). In this project, we aim to explore how Ebola virus GP interacts with TLR4 and determine the effects of TLR4 activation on Ebola virus replication. Intriguingly, the closely related Reston virus does not trigger a TLR4-skewed inflammatory response in human macrophages. Reston virus belongs to the ebolavirus genus but has not yet been associated with human disease. Our working hypothesis is that that the amplitude of the inflammatory response induced by different ebolaviruses correlates with virulence.

induced pluripotent stem cell (iPSC)-derived human hepatocytes. Immortalized cell lines are commonly used to study viral infections. However, these cells do not always recapitulate what is observed in primary cells or organs. Specifically, many immortalized cell lines have blunted type I IFN responses. In collaboration with Dr. Gustavo Mostoslavsky at the Center of Regenerative Medicine (CReM), BU, we have established novel and innovative hepatocyte infection platforms to recapitulate the pathology of filovirus infection in human liver cells. Our data show that there are significant differences in the host response to Ebola virus infection in iPSC-derived hepatocytes cells compared to hepatocarcinoma cell lines.
Antiviral pathways. Filoviruses efficiently block the induction of antiviral pathways in many cell types. We are interested in identifying these pathways and develop tools to reverse the virus-induced block with the goal to boost antiviral responses in the infected cells. Pathways we focus on include the cellular stress response, mRNA regulation, activation of antiviral proteins such as PKR, and pathways that lead to cell death. We are also perform comparative analyses across host species.