Participants
Click on name to see their abstract and elevator pitch video.

Deoxyribonucleic acid, also known as DNA, is a molecule that contains the genetic information in cells. Recent findings suggest that DNA has the ability to conduct electricity, making it a leading candidate for future application in molecular electronics, which could potentially replace current silicon-based technology. However, the electrical conductivity of DNA continues to be controversial due to contradictory results in the literature. To help address these inconsistencies, this work investigates the single molecule conductance and binding of individual DNA component molecules using the Scanning Tunneling Microscope Break Junction (STM-BJ) technique. We seek to identify which chemical moieties in DNA’s nitrogenous bases may bind to the metal electrodes and contribute to the inconsistent conductance signatures of DNA as a whole. Here, we focus on the adenine nucleotide 2’-deoxyadenosine 5’-monophosphate, the associated nucleoside 2’-deoxyadenosine, and adenine itself. We find that adenine and 2’-deoxyadenosine are able to bind in our metal-molecule-metal junctions while the 2’-deoxyadenosine 5’-monophosphate nucleotide does not. Adenine has multiple conductance signatures; based on previous measurements, we conclude that it binds to gold through the imidazole and pyrimidine moieties. In contrast, 2’-deoxyadenosine has only one binding geometry involving the pyrimidine ring. We suspect that steric hindrance prevents the 2’-deoxyadenosine 5’-monophosphate nucleotide from bridging the junction. These results suggest that DNA may be binding directly through multiple nitrogen species on the nucleotides, resulting in inconsistent conductance measurements reported in the literature.

The transcription factor NF-kappaB (NF-κB) is involved in regulating genes that are responsible for immunity and development in humans and has shown to contribute to a variety of human cancers and immune diseases. However, there is still a lack of knowledge about NF-κB in more primitive organisms. To further explore the evolutionary origins of NF-κB, we have been studying predicted NF-κB proteins encoded by single-celled choanoflagellates, especially Acanthoeca spectabilism (As), whose phylum has been suggested to be the closest living phylum to all animals. The As-NF-κB protein sequence is comprised of a highly conserved region known as the Rel Homology Domain (RHD), which is responsible for DNA binding and dimerization in many higher organisms, but does not contain an Ankyrin Repeat domain which is typical of other most other NF-κBs. One of our objectives is to synthesize the NF-κB from A. spectabilism and clone it into a pcDNA-FLAG vector, which contains a FLAG sequence that will be used for detection in biochemical and cellular assays. For that project, we generated an As-NF-κB pcDNA, which was codon-optimized for expression in human cells. We hypothesize that if the choanoflagellate NF-κB gene contains the highly conserved RHD, it will localize to the nucleus to activate transcription, bind DNA, and act like other NF-κB proteins of recently derived organisms. We also mined genomic and transcriptomic data of As to find the NF-κB sequence and compared it to the homology of recently derived organisms as well as other choanoflagellates. Based on this comparative analysis, we were able to identify certain residues, including ones that contact DNA, that are highly conserved across all animal phyla. In future experiments, we will express the plasmid in cells to analyze its ability to bind DNA, activate transcription, enter the nucleus, and be phosphorylated by kinases. These studies will uncover new information about the evolution of the immune system." target="_blank">Click here for elevator pitch
The transcription factor NF-kappaB (NF-κB) is involved in regulating genes that are responsible for immunity and development in humans and has shown to contribute to a variety of human cancers and immune diseases. However, there is still a lack of knowledge about NF-κB in more primitive organisms. To further explore the evolutionary origins of NF-κB, we have been studying predicted NF-κB proteins encoded by single-celled choanoflagellates, especially Acanthoeca spectabilism (As), whose phylum has been suggested to be the closest living phylum to all animals. The As-NF-κB protein sequence is comprised of a highly conserved region known as the Rel Homology Domain (RHD), which is responsible for DNA binding and dimerization in many higher organisms, but does not contain an Ankyrin Repeat domain which is typical of other most other NF-κBs. One of our objectives is to synthesize the NF-κB from A. spectabilism and clone it into a pcDNA-FLAG vector, which contains a FLAG sequence that will be used for detection in biochemical and cellular assays. For that project, we generated an As-NF-κB pcDNA, which was codon-optimized for expression in human cells. We hypothesize that if the choanoflagellate NF-κB gene contains the highly conserved RHD, it will localize to the nucleus to activate transcription, bind DNA, and act like other NF-κB proteins of recently derived organisms. We also mined genomic and transcriptomic data of As to find the NF-κB sequence and compared it to the homology of recently derived organisms as well as other choanoflagellates. Based on this comparative analysis, we were able to identify certain residues, including ones that contact DNA, that are highly conserved across all animal phyla. In future experiments, we will express the plasmid in cells to analyze its ability to bind DNA, activate transcription, enter the nucleus, and be phosphorylated by kinases. These studies will uncover new information about the evolution of the immune system.

Fibrosis is a debilitating condition resulting from dysregulated tissue repair during which fibroblasts produce excess extracellular matrix components, such as collagen and alpha-smooth muscle actin. This response can lead to organ failure, irregular scarring, and severe pain in various contexts, as well as play a role in the growth of the extracellular matrix (ECM) of tumors. A reproductive hormone called relaxin has been shown to abrogate collagen production and increase collagen degradation, thus reversing the underlying cause of fibrosis. This study focuses on the effect of recombinant human relaxin-2 on collagen I production by human dermal fibroblast cells, as dermal fibroblasts play a role in hypertrophic scarring and interact with melanoma cells in skin cancer. This relationship was explored in vitro by culturing human dermal fibroblasts as a monolayer form, in a spheroid form to better represent a three-dimensional environment in vivo, and co-cultured with human melanoma cells to examine the interactions between skin cancer cell growth and dermal fibroblast collagen production. Fibroblasts were stimulated with transforming growth factor beta (TGF-β) to simulate a fibrotic environment and upregulate collagen I expression. Various concentrations of relaxin were added and the resulting collagen levels were observed and compared between the different conditions. Interestingly, both the monolayer and spheroid model show a net decrease of collagen-I with increasing relaxin concentrations. This study demonstrates the potential for relaxin to be utilizes as a therapeutic in the context of skin to reduce fibrotic scarring an ECM buildup by fibroblast interacting with cancer cells.

Citreamicins are a group of natural products with antibiotic and antitumor potential. Isolated in 1989 and 1990 from the bacterial species Micromonospora citrea, these molecules of the polycyclic xanthone family have yet to be synthesized. The Porco Group is working to establish a pathway towards total synthesis: the first step of the proposed process is a silver oxide (Ag2O)-mediated 4 + 2 cycloaddition between a diene and a quinone to form a bicyclic structure. In attempt to make the reaction more successful by increasing yield and efficiency, factors such as concentration, type of solvent, and amount of reagent (Ag2O) were tested. A higher concentration of reactants and reagent and the use of an aprotic polar solvent such as dimethylformamide (DMF) were found to be most advantageous. This finding brings us closer towards the total synthesis of citreamicins, compounds with immense medical potential.

Natural Language Processing is a branch of artificial intelligence that explores how computers can be used to understand and manipulate natural language text or speech. Machine learning models based on different algorithms use texts annotated with labels in order to predict these labels on unseen text. Sentiment analysis is a field within natural language processing where these labels involve opinions expressed in a piece of text, often to determine whether a writer’s attitude towards a certain topic or product is positive, negative, or neutral. In the case of this project, the sentiments to be analyzed are the intentions behind user interactions on a platform for code development known as Github. The goal here is to choose a machine learning model that performs best at classifying peer-review comments on GitHub in three categories: sentiment, hostility, and topic. We are using the built-in models in Python’s sklearn library as the code for our models and MLflow for logging the performance scores. The best performing model is a Logistic Regression model with an average of 73.6% accuracy and .28 seconds training time. The relatively low accuracy is likely due to the little amount of available labelled data. Another working group investigates the level of diversity in the Github community, thus in collaboration with the work here, the combined project will be able to determine the community reaction to contributions from people of different backgrounds.

BPA (bisphenol-A) is a harmful chemical that was once common in plastics in the United States. We have since reduced the amount of this chemical in our plastics, but it still common in other countries. Because of this, it is vital that we understand what BPA is and what its effects are on the human body. In our lab, we use the sea urchin species Lytechinus variegatus as model organisms to determine the effects of varying quantities of BPA on skeletal and morphological development. We investigated which doses of BPA cause a phenotype in the sea urchin embryos and if we are able to pinpoint a specific kind of skeletal or growth defect. We performed dose responses on embryos immediately post-fertilization and then obtained skeletal images of the embryos at 24 and 48 hours post-fertilization. To supplement this, we then performed pre-fertilization dose responses and then obtained images of the fertilized embryos as 1.5 Hours Post-Fertilization (hpf), 22 hpf and 44 hpf. We also performed PMC stains, which show us the locations of the primary mesenchyme cells (PMCs) which secrete the skeleton during varying stages of development. We were then able to see how many were present in the embryos treated with each of the different BPA concentrations, and also where these PMCs were located. From our current data, we predict that there is a range, most likely between 4.5 and 7.5 uM (conc. micromolar) BPA, that causes defects in sea urchin embryos without resulting in death. Our research will be vital in understanding how BPA affects those organisms which consume it, and can contribute to future research in order to mitigate these effects.

Ions colliding onto surfaces under different circumstances result in ultrasmoothening effects or in the formation of nanoscale patterns such as dots and ripples. Understanding the cause and effects is important because ion bombardment is relatively cost efficient than other surface processing techniques, and well-defined structures are essential in electronic and biomedical industries. Our study is focused on the impact of incidence angles on surface stress, which is calculated from wafer curvature measurements collected through the Multi-beam Optical Stress Sensor(MOSS) system, in which laser dots are reflected off of the sample, into a CCD camera, and spacings are recorded. We conducted experiments at angles ranging from 0 to 75 degrees. In the experiments on fresh samples, we find a compressive spike during the beginning of bombardment that is predicted by Swenson’s model and observed in Joy Perkinson's experiments. The peaks of these spikes also decrease in magnitude as incidence angle increases, another agreement with Perkinson’s thesis. Our AFM analysis supports that samples are smoothened under 0 degrees bombardment regardless of previous stress. In the experiments conducted on pre-smoothened samples, we find a small and brief compressive spike followed by tensile stress at all angles, which is not predicted. Both types of experiments, however, do not show signs of Norris’ model becoming dominant after initial swelling as Perkinson speculated.

Two dimensional MXenes have been a broad focus of study in recent years due to their emerging properties and importance in the materials science industry. The conversion of tungsten disulfide to tungsten carbide is an important research topic because of the practical uses of tungsten carbide.The optimal conditions for this conversion process to occur are being analyzed. Changing temperature and gas flows will be analyzed to see the effects on the efficiency of the reaction to identify the most effective conditions for the reaction to occur. Through the conversion process, gas flows will be introduced to the sample to optimize conditions. Optical images taken before and after suggest optical changes. Samples are further characterized using Raman spectroscopy to analyze the samples before and after to see the change from WS2 peaks to WC peaks. Conversion conditions were further optimized and Raman was compared between samples. Atomic force microscopy is measured to ensure good crystallinity, identify the thickness changes, and the smoothness of the surface. Transmission electron microscopy confirms the crystal structure of the synthesized material. The practical uses of these materials are for electronic devices and memory storage devices. In order to create a functional device, these materials must be effectively converted

Increased nitrogen loading caused by fertilizer runoff and wastewater have significantly increased the production of phytoplankton and microalgal biomass, leading to problems of eutrophication at Waquoit Bay. As a result, greater amounts of microphytobenthos have been observed. This decreases oxygen levels, which leads to the death of vital plants and organisms, altering the biology and chemistry of the marine ecosystem. Chlorophyll-a produces oxygen through photosynthesis, and as a result, can be used to understand oxygen levels. The objective of this study was to observe the relationship between sediment chlorophyll-a (Chl-a) concentrations and oxygen (O2) profiles from sediment cores at three sites in Waquoit Bay. Sediment oxygen and temperature profiles were measured in 0.5 cm increments up to 2 cm and in 1 cm increments up to 5 cm using a fiber-optic oxygen and temperature sensor by Pyroscience. In addition, we extracted sediment chlorophyll-a and water column chlorophyll-a and measured their concentrations using a fluorometer. Hach probes were used to characterize the environmental parameters of the surface and bottom waters, including pH, salinity, oxygen and temperature. Overall, our results showed that oxygen levels decreased as depth increased at all sites. However, O2 concentrations were slightly higher at the Head of the Bay where the sediment was sandy, while the two muddier sites (BW2 and SWMP) quickly declined in oxygen within 1.0-1.5 cm. This may be because sand is porous. In addition, our data showed that sediment chlorophyll-a concentrations positively correlated with oxygen levels at each site. Since chlorophyll-a produces oxygen, areas with more chlorophyll-a would have more oxygen. Through all of this, we have a greater understanding of the variables that factor into oxygen levels, which is essential for the preservation of organisms and plants in the marine ecosystem.

Spatial Frequency Domain Imaging (SFDI) is a diffuse optical imaging technology that provides a non-invasive method to measure metabolic and molecular features of skin tissue, which provides a new means to track treatment response and chemotherapy resistance of tumors in women with breast cancer. SFDI uses diffuse reflectance measurements of spatially modulated light to provide quantitative measurements of the optical absorption and reduced scattering coefficients of tissue, allowing medical professionals to monitor metabolic information such as tissue oxygenation and chromophore concentration (e.g. oxyhemoglobin and deoxyhemoglobin) of a breast cancer patient in clinical applications. Currently, there has been no research performed on the accuracy and precision of the SFDI system device being developed in Professor Robyler’s BOTLAB, so there are uncertainties concerning the reliability of the device’s data. We aim to compare the data of the BOTLAB’s SFDI system to a commercial system to quantify the accuracy and precision of the BOTLAB’s SFDI system. Our findings will allow us to correct spatial frequency collection and further develop the SFDI system for future applications in cancer research.

Neurogenesis is the birth of new neurons. Humans lose this ability early in most of the brain, while other species including songbirds retain neurogenesis for their entire adult lifespan. This research project aims to address how neuron migration, a key stage of neurogenesis, varies with behavioral state (e.g. sleep) in the zebra finch. We examined two methods of imaging neuron migration: single-photon or two-photon microscopes in wild type or transgenic zebra finches. Here, we present data demonstrating the strengths and weaknesses of both approaches.
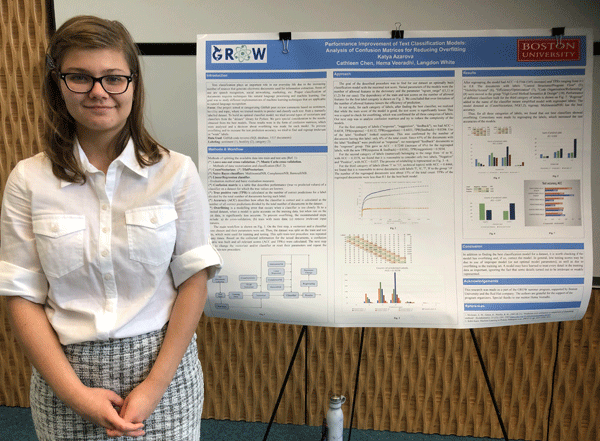
Text classification plays an important role in our everyday life due to the increasing number of sources that generate electronic documents used for information extraction. Our project was on text category analysis (sentiment, hostility, topic), where we created a program that predicts labels for a manually labelled dataset. Our goal was to study different implementations of machine learning techniques that are applicable to natural language recognition and methods of improvement of recognition accuracy. To build an optimal classification model, we used two cross-validation methods and tried several vectorizers and classifiers. The main workflow was as follows. On the first step, a vectorizer and a classifier were chosen and their parameters were set. Then, the dataset was split on the train and test sets. This split-train-test procedure was repeated many times and scores were calculated and stored. The next step was to change vectorizer and/or classifier or reset their parameters and repeat the split-train-test procedure. The goal of this procedure was to find a classification model with a maximal test score. We gave special consideration to the results obtained from the best models. They were in the form of confusion matrices, which were analyzed and a decision about overfitting was made for each model. Overfitting is a modeling error that occurs when a classifier is too closely fit to a limited dataset, when a model is quite accurate on the training data and significantly less accurate on the test data. To prevent overfitting and to increase the test prediction accuracy, we tried to find and regroup irrelevant or "weak" labels. In our study, for each category of labels, after finding the best classifier, we checked overfitting, which was confirmed for all categories. By analyzing confusion matrices we were able to reduce the complexity of the models by regrouping the labels, which increased the test accuracies of the models.

Quantum dots are semiconductor nanocrystals that can be used in biosensors as fluorescent labels due to their stability and brightness. Here, code in MATLAB was developed to perform automatic image processing as an alternative to manual analysis in ImageJ. The goal was to reduce error, improve efficiency, and maintain consistency when analyzing smartphone images of quantum dot bioassays. The algorithm was designed to threshold, crop, and select specific regions of interest in each sample in the images. Each region of interest was analyzed in the red channel and the average pixel intensities and standard deviations were calculated. The code was tested across multiple images to ensure consistency. The coefficients of variation of the final response curves were consistently lower when the images were analyzed by the code, compared to the same images analyzed in ImageJ. Additionally, the time necessary to analyze the images were significantly reduced by the use of the code. In the future, the improved efficiency from this code will be implemented in a Raspberry Pi to produce immediate readouts of the quantum dot bioassays, furthering the development of these assays for clinical use in a point-of-care diagnostic tool.

Programmed cell death is a necessary process in development and in many disease pathways, such as in neurodegeneration, cancer, and asthma. There are various forms of programmed cell death, and while the most well-known type of cell death, apoptosis, has been well studied and characterized, much about the processes of other, non-apoptotic forms of cell death remain unknown. During oogenesis in the Drosophila ovary, nurse cells are murdered and cleared away by follicle cells. Through studying the role of various proteins in programmed cell death we can better understand its mechanisms. Recently, the McCall lab has conducted a proteomics analysis to determine the secretome, the proteins being secreted by the endoplasmic reticulum of follicle cells in the Drosophila ovary. We aim to further investigate the role of these proteins through knocking them down to observe their effects. To validate whether these secreted and transmembrane proteins are involved in nurse cell death, we performed an RNAi screen to knock down genes that code for these proteins. Once the genes were knocked down, we used DAPI to stain DNA to observe any abnormalities in cell clearance. Through antibody staining of cell membranes, we can observe abnormalities in engulfment. RNAi knockdowns of these proteins did not have a significant impact on late-stage cell death. However, loss of function of Kruppel homolog-2, a transmembrane protein, and thick veins, a receptor for a growth factor regulating survival and death signals, in the follicle cells results in mid-stage nurse cell death, indicating that these proteins are involved with the prevention of mid-stage cell death.

Connections between neurons form both large and small scale brain circuits which have diverse functions. Current techniques lack the ability to record neural activity at a high spatial resolution with cell type specificity over a large volume of brain tissue, which is necessary for fully understanding the signaling dynamics within these brain circuits. To address this, we developed high density fiber photometry arrays, which are 3D printed grids holding over one hundred custom oriented optical fibers that transmit fluorescence. To pilot this technique we studied midbrain dopaminergic neuronal projections to the striatum in behaving mice receiving unpredicted rewards and spontaneously running on a spherical treadmill. We created high-density fiber arrays that would capture the full functional topography of signaling across the entire striatum and then implanted these arrays in the striatum of DAT-cre mice that were virally injected in the dopaminergic regions of the midbrain with flexed GCaMP6f, a calcium indicator. We used single photon excitation and imaging through a CMOS camera to successfully record changes in fluorescence through the fibers, indicating active signaling from the dopamine axon terminals. These optical measurements demonstrated functionally heterogeneous dopamine signals present throughout the striatum. Further analyses will investigate the 3D topography of these signals on multiple spatial (100s of microns to millimeters) and temporal (10s of milliseconds to seconds) scales. High-density fiber photometry will be widely applicable to studying relationships between behavior and distributed cell-type specific neural signaling across the brain.

Toughness, defined as the energy absorbed prior to failure, plays a critical role in the design of failure resistant materials and structures. Designing for high toughness is challenging because it requires a balance of both strength and ductility, two properties which tend to be mutually exclusive. This challenge is further compounded when considering the stochastic nature of failure and the difficulty in reliably predicting the influence of defects and manufacturing variability. As a result, designing structures for high toughness requires numerous experiments to explore the design space. The high level of control afforded by recent advancements in additive manufacturing (AM), commonly known as 3D printing, enable opportunities to rapidly design and test structures. In this work, we utilize AM to explore the toughness of 3D printed hollow cylinders under quasistatic compression. The hollow cylinders are 3D printed from polylactic acid (PLA) and tested on a universal testing machine (5965 Instron Inc.). By fabricating and testing hollow cylinders of varying heights, thicknesses, and diameters, we were able to identify the characteristics of a hollow cylinder with high toughness. From our findings, we also observed that cylinders exhibiting shell buckling possessed high toughness and cylinders exhibiting Euler buckling possessed low toughness. Interestingly, the PLA hollow cylinders depicted similar buckling behaviour to studies using aluminum hollow cylinders. While in our work we investigated the toughness of a simple hollow cylinder, the design freedom afforded by AM opens new opportunities to explore more complex designs.

Rhodamine is a fluorescent dye that spontaneously converts between a bright and dark state at physiological conditions, emitting light at different wavelengths depending on the molecules that are conjugated to it. Therefore, it can serve as a good fluorescent probe for identifying biological targets. This study investigates the interactions between rhodamine dye derivatives and amino acids, and attempts to synthesize an optimal dye that will emit a specific max wavelength for each amino acid. The 20 amino acids are each conjugated to different rhodamine dye derivatives. These dyes were synthesized using a divergent synthesis route which allowed us to obtain a library of derivatives to test for the desired properties. The results show that when the dye is not conjugated to any amino acid, it emits max wavelength around 583 nanometers; when the dye is conjugated to amino acids, the max wavelength emitted exhibit shift patterns specific to each amino acid. While doing experimental testing, calculations were also done using a computer software called Maestro to rationally design a derivative that will provide the most direct amino acid-dye interactions. The results of my work will help scientists to image polypeptides under high resolution, thereby developing a new assay to further investigate proteins and their cellular localization, which can be used to potentially determine new targets for treating diseases such as cancer.

For some legal reasons, I cannot disclose the entirety of the project I worked on. However, I can discuss some of the programming that I did. This summer, in collaboration with GROW, Red Hat, and BU Spark!, I had the opportunity to analyze and migrate some data about baseball umpires in order to better fit a baseball research project. Working alongside another backend developer, I helped developed python scripts that analyzed this data and reorganize it to fit the needs of research. In these last six weeks, I’ve been programming in python. A part of my project was working on merging two separate datasets together to make a combined dataset. In computer science, these organized datasets are called CSVs. CSV stands for “comma separated values”. These large files of values are hard to read and use without organizing it in a way that makes it easier to use. I wrote a script that accomplished this. This program merged the two sets of data (CSVs) together and also converted the final product into a much easier to use format called JSON. JSON files are similar to tables, but they can be read as a standard text file. The python script I wrote sorted the data by name, so that the umpire information would be easier to understand for the researchers.The secondary part of the project I was working on is updating the information on the umpires. Umpire data changes relatively quickly, so it’s important to update it constantly. In order to do this, I combined the two programs together so that there could be automatic updates to the already translated files as soon as a new piece of information was placed in the raw files.I accomplished this by merging the two programs together, and then changing what input they take in, which reduced the difficulty of integrating them together. When these programs were separated, each had to be called separately in order to complete the task at hand. One was for uploading the umpire information, and the other is for refreshing the umpire information. With this new merging approach, one program would be able to do each part simultaneously. In addition to this work, I also worked on merging large amounts of data together using a python script I wrote. Because the research being conducted collects data on umpires over several years, there is a lot of raw data to work with. Again, this data is in CSV format, which is hard to read and understand. Also, there were several folders sorted by year that contained multiple large CSV files with data. To make the data simpler for the researchers, I wrote a script that compiled all of these files together. By having the script go through each CSV line by line, and matching the name of the umpire to the corresponding line in a secondary CSV, I was able to start the merging process. After the match was found, I collected that data and added it to a new file. I continued this process until all CSV’s in that particular folder had been sorted through. Once all umpire information was in a file, I converted it from CSV to JSON, making it easier for future programmers and researchers to work with.

Skeletal muscle is integral to daily life. While it is normally capable of regeneration, there are instances of debilitating muscle loss—as seen in dystrophic diseases and during aging—where regenerative capacity is exhausted. Despite advances in identifying genes involved in regeneration, the molecular mechanisms of this process have not been fully defined. Our lab is investigating a regulator in this process which might better inform muscle regeneration strategies in the future. Long noncoding RNAs (lncRNAs) have emerged as key regulators of gene expression. Preliminary data from our lab suggests that one lncRNA in particular, Meg3, is required for normal muscle regeneration to occur. Meg3 was knocked down in injured skeletal muscle using an adenovirus-mediated short hairpin RNA (shRNA), which resulted in impaired muscle regeneration. However, the regenerating muscle environment has many different kinds of cells in addition to muscle, including neurons, stem cells, white blood cells, as well as fibrotic cells. It remains unclear which specific types were being affected by the knockdown Meg3 and thus altering the regeneration process. For this project, mouse hindlimb muscle was injected with cardiotoxin to trigger muscle degeneration followed by regeneration. Co-injection of a virus overexpressing β-galactosidase was used to identify those cells which were transduced, i.e. infected, by the virus. To characterize the types of cells preferentially transduced by adenovirus, a β-gal assay and immunostaining were performed. Three days after injury hindlimb muscle was isolated, cryosectioned, and subjected to the aforementioned assays. The samples were subsequently imaged and analyzed using ImageJ. Immunofluorescence labeling revealed that regenerating myofibers were preferentially transduced by adenovirus, which suggests that adenovirus-mediated Meg3 knockdown primarily occurred in muscle cells. Further investigation is required to determine the precise mechanisms of regenerative defects associated with Meg3 knockdown. This will not only broaden our understanding of muscle regeneration, but also lead to improved strategies towards restoring muscle in patients who desperately need it.
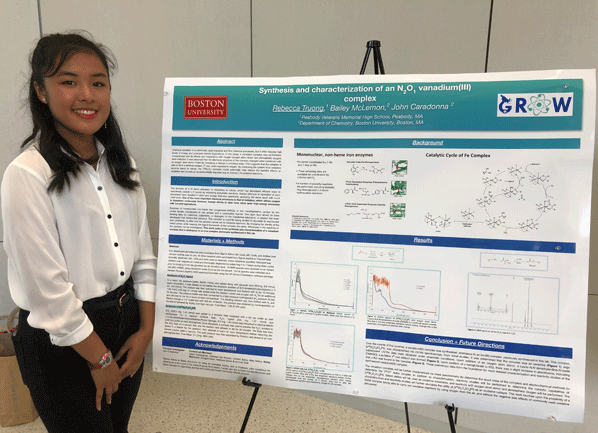
Chemical oxidation is a commonly used industrial and fine chemical processes, but it often requires high levels of energy and produces harmful byproducts. In this study, a vanadium complex was synthesized, characterized and its effects and interactions with oxygen (oxygen atom donor and atmospheric oxygen) were analyzed. It was observed that the structure of the complex changed when combined with an oxygen atom donor, indicating a change in oxidation states. This suggests that the complex is able to form a terminal oxidant, VV-oxo, when exposed to oxygen. By mimicking the system of an oxidative enzyme found in nature, the VIII(N2O1) complex could potentially help reduce the harmful effects of oxidation and provide an environmentally friendlier way to induce oxidation.

An autocorrelator is used to measure the duration of ultrashort pulses, which can have a time duration in the femtosecond (10-15 of a second) range or shorter. Other common devices used to measure pulse duration cannot be used in this instance because they are not accurate on the femtosecond time scale and would be used for longer pulses. An autocorrelator works by measuring the pulse against an identical copy of itself, one of which is delayed in order to cause the two pulses to collide in different places. This collision occurs inside a nonlinear crystal, which releases a third pulse, with the intensity of the two pulses combined. The intensity versus delay are plotted and the time duration is found with the full width half maximum (the width between the two points that are half of the maximum point of the function). Here, we demonstrate the steps to program the delay stage of the autocorrelator which is used to delay the second pulse. In order to accomplish this task, we used LabVIEW which is a systems engineering software that uses a visual programming language. Using LabVIEW, the delay stage of the autocorrelator can be moved according to an expected pulse duration which allows for the intensity to be measured at different delays. The time duration of ultrashort laser pulse provides necessary information for further experiments and calculations. Ultrafast laser pulses can help scientists see processes that take place on the femtosecond time-scale, such as chemical reactions and molecular dynamics.

Recent studies have shown that the estrogen steroid hormone 17-β-estradiol has been entering aquatic ecosystems due to industrial activities and human sewage. In excess, 17-β-estradiol and endocrine disrupting hormones such as bisphenol A (BPA) can lead to hormonal perturbation in humans, especially in the development of the reproductive system. Previous work in our lab has shown that BPA, an estrogen mimicker, causes skeletal patterning defects in Lytechinus variegatus larvae. Our investigation seeks to explore how 17-β-estradiol perturbs skeletal development in L.variegatus. In this study, we tested a range of environmentally relevant doses of 17-β-estradiol between 50 ng/mL and 450 ng/mL to identify concentrations that produced developmental abnormalities. At intermediate doses, we observed rotational defects, spurious elements, and deletions in skeletal development. The primary mesenchyme cells (PMCs) which control skeletal development were not perturbed at an early stage suggesting that estradiol takes effect at a later stage. Doses higher than 250 ng/mL were lethal, as most embryos died before the pluteus stage. My work can be used to predict the potential effects of continuous sewage leaks in marine environments and help inform governments seeking to regulate the concentrations of chemicals in ocean water to reduce human exposure to 17-β-estradiol.

In this study, we are analyzing the microbiome of Porites lobata - coral found in Fouha Bay, Guam - and the sediment in their surroundings by using both the coral’s and the sediment’s DNA. Fouha Bay is a region where the Fu Sa river flows into the ocean, carrying fresh water and sediment into the ocean. The sediment can be harmful to the corals that live along Fouha Bay because it prevents coral from performing photosynthesis and increase the risk of coral disease. The purpose of the research is to examine the bacterial community that lives within the coral and its surrounding environment, specifically across the sediment gradient that exists within the bay. We are investigating if there are similarities between the coral microbiome and the bacterial community found in nearby sediment as well as how this changes based on where we same on the coral colony. We used 16S and ITS2 DNA metabarcoding to characterize the coral microbiome and the bacterial community of the sediment surrounding the coral colonies. The impact of these results can help scientists understand the role the coral microbiome plays in coral health and how it responds to different environments.