Target Discovery & Proteomics Lab
Drug discovery is inherently risky and extremely costly; it takes years for compounds to reach the clinic and market to recoup the large investments made. Biological target identification and validation (“target ID”) has emerged as a critical challenge in the drug discovery pipeline and involves the identification of one or several proteins from among the many thousands present in cells or tissues that are responsible for biological effects of a bioactive compound. Target ID is therefore essential for understanding a lead compound’s mechanism-of-action, adds tremendous value to promising chemical probes and therapeutic leads, and allows poor lead compounds to fail earlier. However, target ID remains a major unaddressed challenge for many research programs in academic labs and in the pharmaceutical/biotechnology industries.
The BU Target Discovery and Proteomics Lab (BU-TDPL) was founded in 2021 by BU-CMD Director Professor John Porco with the aim of resolving a key bottleneck in drug discovery: inadequate target ID. To address this challenge, the TDPL has implemented innovative compound-protein interaction mapping technologies based on precision mass spectrometry that allow for systematic identification of the protein targets of bioactive leads from compound screens to characterize mode-of-action and structure-activity relationships. This laboratory is currently housed on the Charles River Campus (LSE 712) and is closely interfaced with the BU Center for Molecular Discovery (BU-CMD) on the CRC, and chemical/biological investigators on both the CRC and medical campuses.
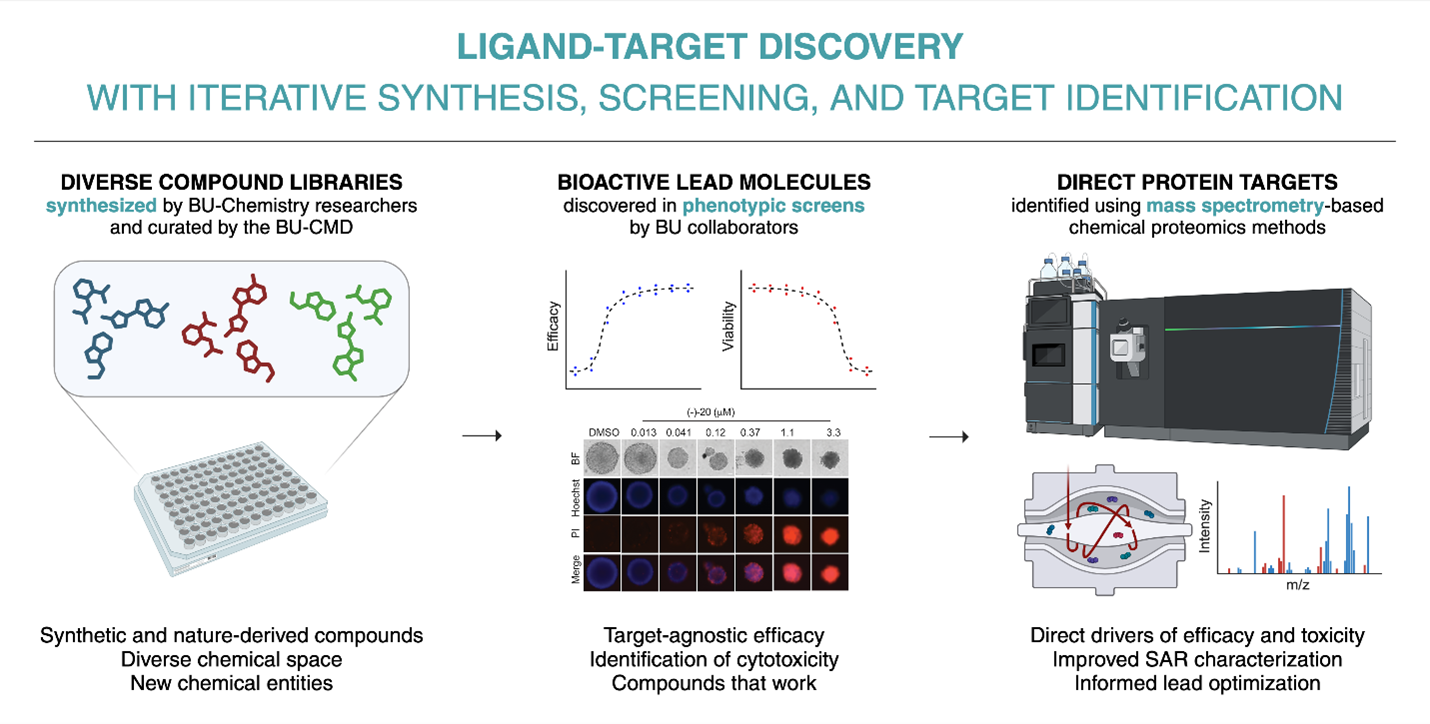
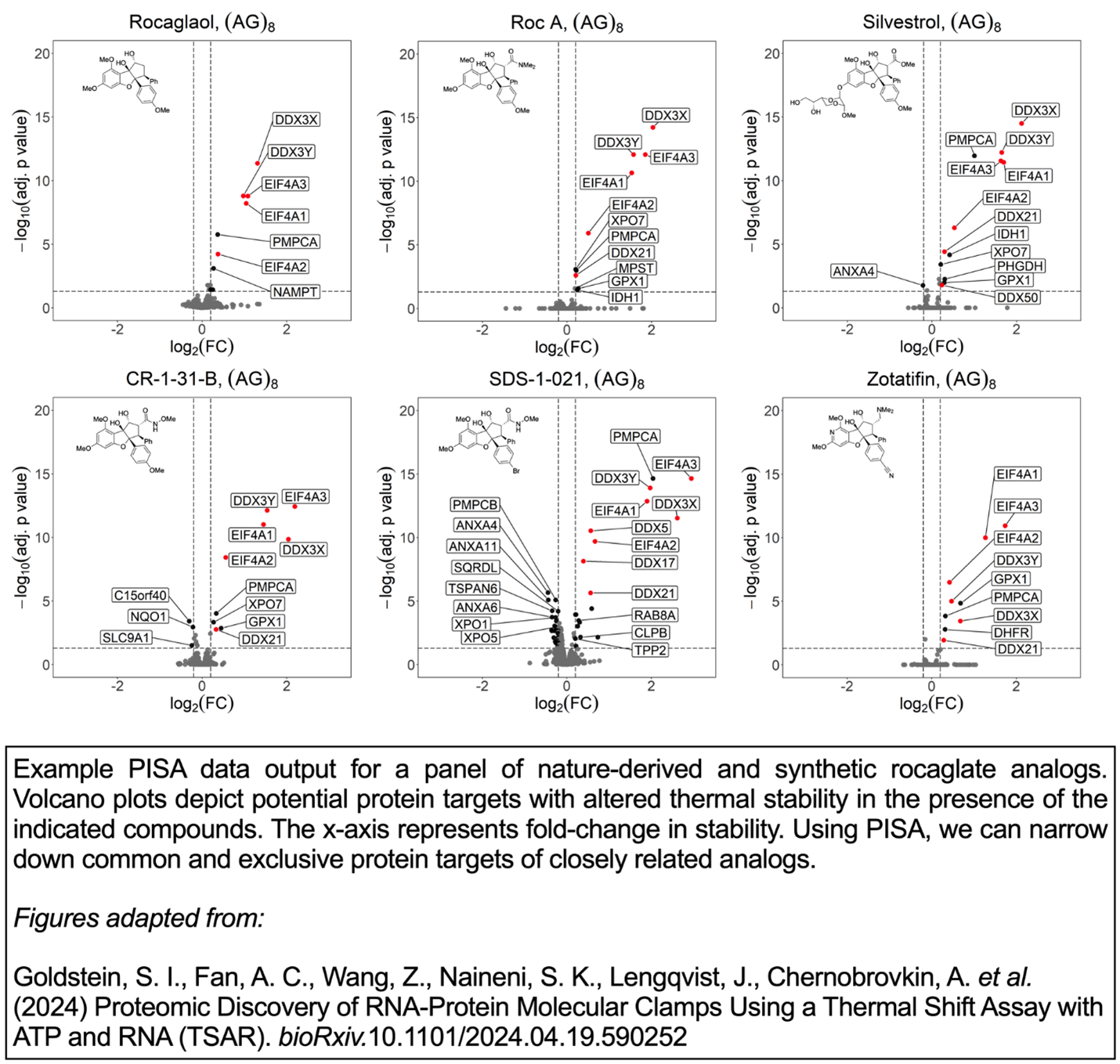
One core capability of the BU-TDPL is the Proteome Integral Solubility Alteration (PISA) assay (1), a variation of the mass spectrometry-based Cellular Thermal Shift Assay (CETSA®) (2) developed by Pelago Bioscience which enables untargeted detection of small molecule-protein interactions on a proteome-wide scale. The PISA assay exploits the fact that, whereas most cellular proteins tend to aggregate upon heat denaturation, ligand-bound proteins display enhanced thermal stability and are less prone to precipitate after heating. Using an Orbitrap Eclipse Tribrid Mass Spectrometer with FAIMS obtained in 2020 under a High-End Instrumentation (HEI) grant proposal (1S10OD026807, “Ultra high precision mass spectrometer”), we can measure ligand-induced protein stability shifts in a systematic quantitative manner to identify the targets of bioactive compounds in different biological contexts. An example data output from our published PISA experiments is shown below.
The BU-TDPL has also started to gain traction in funding from research proposals, including a recent research award from the BU CTSI Pilot Grant Program (1UL1TR001430-Target Deconvolution of Host-Directed Antiviral Rocaglates). Additionally, we have reached a collaborative agreement with Pelago Bioscience AB, holders of the CETSA® patent family, to use PISA in academic research projects.
Moving beyond CETSA®-based methods, the BU-TDPL now employs a broader selection of proteome-wide target ID techniques including quantitative affinity enrichment experiments with immobilized small molecules (e.g. biotinylated probe pull-downs), a historic workhorse in the target ID toolkit. In the future, fee-for-service proteomics experiments such as molecular weight determination, protein identification, quantitative analysis (both multiplexed and label-free assays), post-translational modification analysis, and more will be available. Fit-for-purpose and novel target ID method development, including affinity enrichment with custom probes, project-specific method optimization, and new BU-TDPL research endeavors will keep the Center on the cutting edge of technological innovation.
