Analysis of ligand binding to DNA and RNA molecules
Vajda, Kozakov, and Frank-Kamenetskii groups
Small molecules that bind to DNA are extremely useful as biochemical tools for the visualization of DNA both in vitro and inside the cell. Additionally, the clinical significance of DNA-binding compounds can hardly be overstated, as many anticancer regimens include a compound that binds to and/or modifies DNA. Similarly, RNA is increasingly considered as drug target. Technical progress in RNA synthesis and structure determination is fueling a rapidly expanding knowledge about RNA three-dimensional structure and its roles for biological functions and is paving the way for rational design of therapeutic compounds which bind specifically to RNA folds.
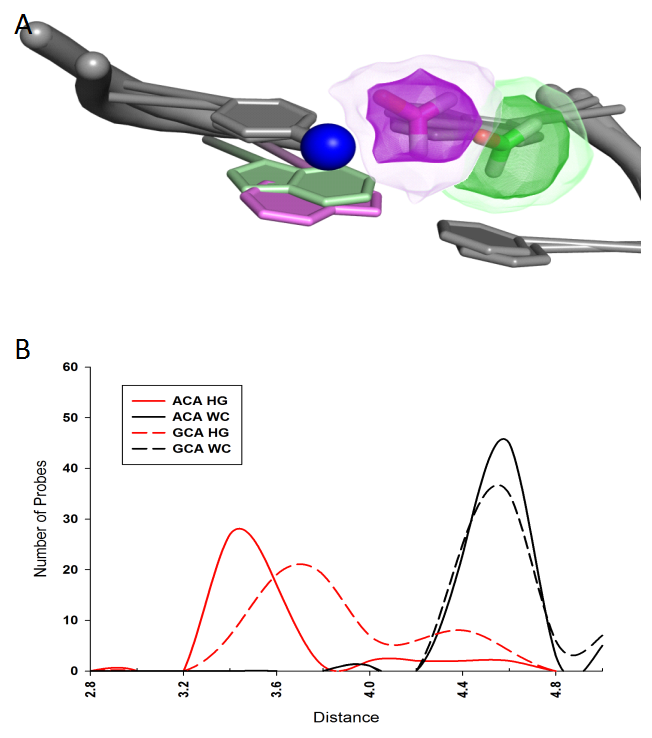
We have studied the interaction between DNA and formaldehyde, which has long been recognized as a hazardous environmental agent highly reactive with DNA. Recently it has been realized that, due to the activity of histone demethylation enzymes within the cell nucleus, formaldehyde is produced endogenously, in direct vicinity of genomic DNA. Should it lead to extensive DNA damage? Previous models did not explain the origin of reactivity of duplex DNA with respect to formaldehyde. Although it was shown that fluctuational openings of base pairs play an important role in the process, there was also a strong indication that a major reaction, hydoxymethylation of the cytosine amino group, proceeds without full base pair openings. We addressed this problem by extending the computational solvent mapping method to nucleic acids. We focused on the leading reaction of formaldehyde with free bases: hydroxymethylation of cytosine amino groups. Our results showed that in B-DNA cytosine amino groups are totally inaccessible for the formaldehyde attack. Then we explored the effect of recently discovered transient flipping of Watson-Crick (WC) pairs into Hoogsteen (HG) pairs (“Hoogsteen breathing”), and showed that the HG base pair formation dramatically affects the accessibility for formaldehyde of cytosine amino nitrogens within WC base pairs adjacent to HG base pairs. The analysis emphasized the significance of DNA HG breathing. Fig. 1 shows the distribution of formaldehyde positioning near the amino nitrogen atom of cytosine. (A) Superimposed formaldehyde (sticks) positioning near the amino nitrogen atom (blue sphere) of cytosine in DNA with Hoogsteen pairing (purple) versus Watson-Crick pairing (green) at the adjacent base pair. The density map shows distribution of low energy formaldehyde positions. (B) Distributions of formaldehyde positions around the amino nitrogen atom for DNA with HG and WC pairing. According to these figures, formaldehyde interacts much stronger with DNA if it has HG rather than WC pairing.
Our recent collaboration focuses on RNA-protein and RNA-drug interactions. RNA structures ubiquitously undergo conformational rearrangement upon complex formation with protein targets and drug molecules. These rearrangements frequently involve large reorientation of intact secondary structural elements about key recognition sites, and made it questionable whether RNA represents a good drug target. However, the analysis of multiple RNA conformers using computational solvent mapping has shown that major binding hot spots remain invariant in spite of the large conformational changes. In particular, we study the class of compounds that have the ability to bind strongly to HIV-1 TAR RNA. The TAR is an RNA domain found at the 5’-end of all pre-messenger RNA transcripts. The interaction between TAR and the trans-activator protein Tat is essential for HIV-1 viral replication, and has been studied extensively as a potential target for anti-HIV intervention therapy.