News
Lynne Chantranupong Receives 2025 Sloan Research Fellowship

Dr. Lynne Chantranupong recently received the 2025 Sloan Research Fellowship. The Alfred P. Sloan Foundation recognizes standout young scientists whose work is promising and innovative. The winners each receive $75,000 over two years.
When Dr. Chantranupong joined the BU's faculty in January 2024, she launched multiple research projects to investigate cellular mechanisms in the brain. One of her focuses is elucidating how a specialized part of the cell, called the lysosome, gets rid of waste, and how this waste differs across neurons.
She recently started collaborating with Dr. Ji-Xin Cheng to image the contents of a single lysosome at higher resolutions. The Sloan Foundation award will support their collaboration and bolster Dr. Chantranupong’s ongoing research projects.
Read the full announcement here.
Congratulations, Dr. Chantranupong!
Five MCBB Faculty Receive 2024 Rajen Kilachand Fund for Integrated Life Sciences & Engineering Awards
Five MCBB Faculty are part of three groups to win 2024 Rajen Kilachand Fund for Integrated Life Sciences & Engineering Awards. The Fund, launched in 2017, has awarded $14 million to support projects that have advanced science, built collaborative structures for interdisciplinary research, and expanded funding opportunities. Learn about their projects below.

Dr. Wilson Wong and Dr. Florian Douam (BU School of Medicine), along with Dr. Alexander Green, Dr. Mark Grinstaff (BU College of Engineering), and Dr. Ahmad Khalil hope to to pioneer a new tool for fighting future pandemics.
About a year ago, Dr. Wong and Dr. Grinstaff discovered a novel way to modify saRNA, a breakthrough they’ve since used to create a more effective COVID-19 vaccine in collaboration with Dr. Douam. Combining this breakthrough with Dr. Khalil’s nanobody discovery platform and computational tools designed by Dr. Green, the team aims to create scalable, cost-effective, and rapidly deployable solutions for viral prevention and treatment.
Dr. Wong is leading the project with Dr. Douam; Dr. Green, Dr. Grinstaff, and Dr. Khalil are coinvestigators.
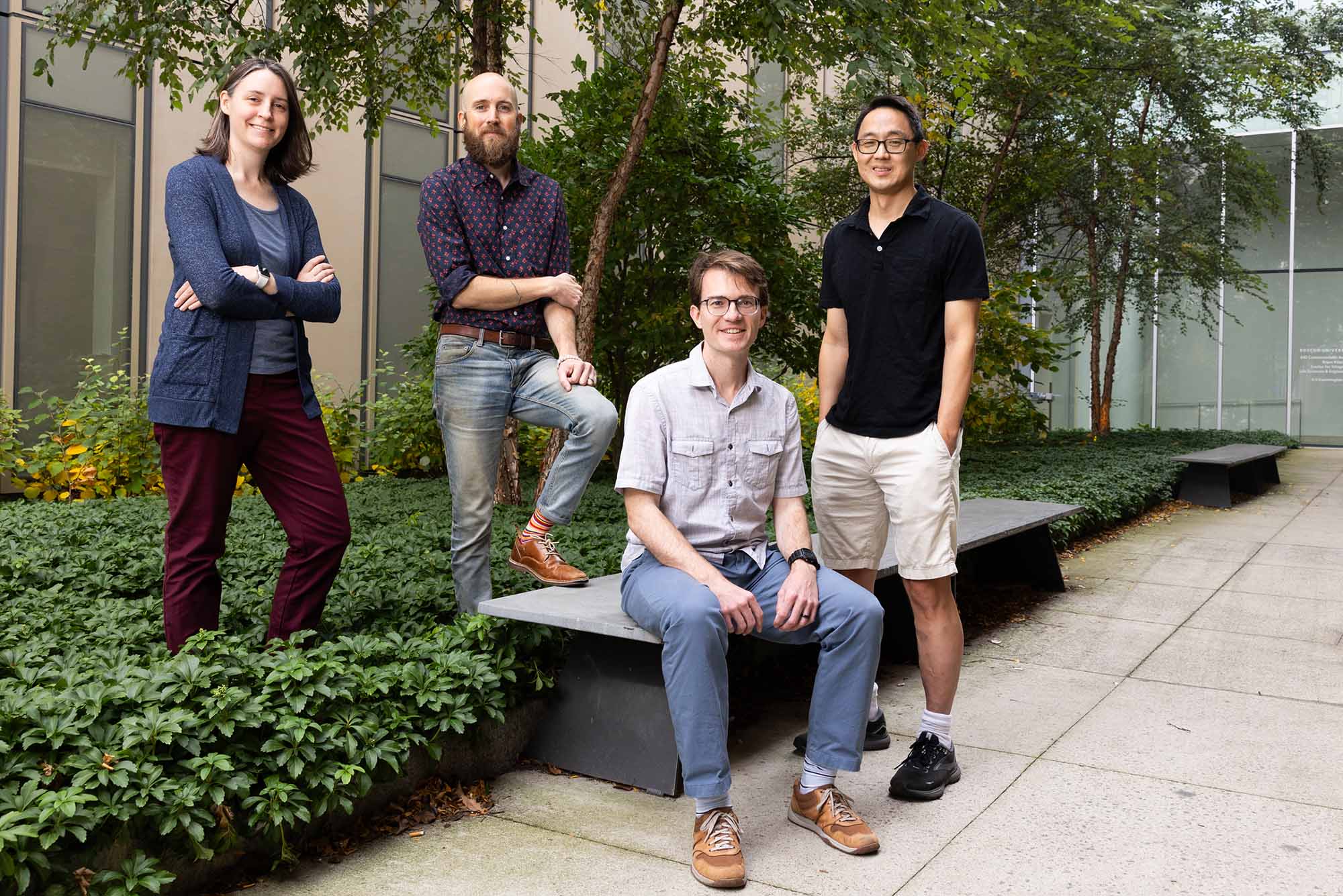
Dr. Joe Larkin and Dr. Mary Dunlop, along with Dr. Douglas Holmes and Dr. Harold Park, are leading innovative efforts to address the urgent global challenge of antibiotic resistance. Antibiotic resistance poses a serious global health threat, with bacteria evolving faster than new drugs can be developed.
The team's project focuses on horizontal gene transfer and the single-cell level events that are involved in making an individual bacterium transition from being drug-susceptible to drug-resistant. Using optogenetics, they aim to trigger and observe this process in real time, offering novel insights into how bacteria transition from drug-susceptible to drug-resistant. Their innovative work could lead to groundbreaking strategies to combat the spread of antibiotic resistance.
Read the full announcement here.
Congratulations, everyone!
Dr. Amanda Pinheiro Published in the Journal Development
 Dr. Amanda Pinheiro, an MCBB PhD alum of the Naya Lab, recently published a paper in the journal Development.
Dr. Amanda Pinheiro, an MCBB PhD alum of the Naya Lab, recently published a paper in the journal Development.
Dr. Pinheiro's paper was selected to appear as a “Research Highlight”, and both she and Dr. Naya were interviewed by the journal in “The people behind the papers” to discuss the significance of their work. This research article, “The Dlk1-Dio3 noncoding RNA cluster coordinately regulates mitochondrial respiration and chromatin structure to establish proper cell state for muscle differentiation”, demonstrates that these RNAs work together to regulate metabolism and epigenetics in muscle. You can read the paper here.
Congratulations Mandy!
Dr. Alexander Green Receives 2024 National Institutes of Health Director’s Transformative Research Award

Dr. Alexander Green, an Associate Professor for Biomedical Engineering in the Green Lab, received the 2024 National Institutes of Health Director’s Transformative Research Award.
The Transformative Research Award focuses on significance and innovativeness where the proposed project must address a significant problem and be bold, highly innovative, and transformative. The proposal must be high-risk, high-reward, and would not be funded through a conventional NIH funding mechanism. Lastly, the application will be anonymized to be reviewed by the NIH staff.
In Green's research, he hopes that by tracking the forces involved when cells interact with one another, like in cell-based cancer therapies, he can better control their behavior and pave the way for more potent disease treatments. It’s an out-there, novel idea—and it could fizzle or it could change lives. But Green’s chances of success have just been given a major boost.
Green is sharing the award’s $7.2 million funding with two researchers from Yale University, Julien Berro, and Xiaolei Su, and he says their proposed cell forces project would typically be a tough one to win backing for—all promise with, so far, little proof that it’ll work.
Green is just the third BU researcher to be given the honor, behind Bela Suki, an ENG professor of biomedical engineering, and Steve Ramirez (CAS’10), a College of Arts & Sciences associate professor of psychological and brain sciences.
Read more here.
Congratulations Dr. Alexander Green!
Dr. Brian Cleary Receives Career Development Professorship

Excerpt from The Brink "Career Development Professorships Awarded to Five BU Researchers"
They’re scholars of Latin American literature and Asian history, experts in biomedical engineering and computing, organic chemistry, and biology—and now, five rising star Boston University faculty are also winners of prestigious Career Development Professorships. The high-caliber awards, given annually by BU’s Office of the Provost, recognizes “talented junior educators emerging as future leaders within their respective fields.”
The professorships provide stipends for the recipients’ salaries and research for the next three years. The awards are named in honor of the donors and alumni who fund them, with winners nominated by their respective deans and selected by the provost’s office.
“Our current cohort of Career Development Professors are incredibly impressive both in terms of the breadth of their work and the impact they have already had on their fields. They bring energy, exciting new ideas, and fresh approaches to their research and teaching,” says Gloria Waters, University provost and chief academic officer. “We are grateful to our generous donors and alumni for their support and shared belief in the promise these rising colleagues present.”
This year’s Career Development Professors are:
Shibulal Family Career Development Professorship, established by BU Trustee S. D. Shibulal (MET’88)
Brian Cleary, a Faculty of Computing & Data Sciences assistant professor, who holds appointments in biology and biomedical engineering, investigates the nature of molecular, cellular, and tissue processes by integrating computational and algorithmic tools into biology research. Called “a deep thinker and fearless leader-in-the-making,” by his nominator, Azer Bestavros, a William Fairfield Warren Distinguished Professor of Computer Science and associate provost for BU’s Faculty of Computing & Data Sciences, Cleary “truly embraces the integrative, cross-cutting culture of CDS and embodies the qualities needed to be a leading innovator in data science.”
Read more here.
Congratulations Dr. Brian Cleary!
Dr. Mary Dunlop Receives New BU Postdoctoral Awards Excellence in Mentorship and Research

Excerpt from The Brink "New BU Postdoctoral Awards Celebrate Excellence in Mentorship and Research"
When postdoctoral researcher Jean-Baptiste Lugagne began applying to faculty positions last year, he steeled himself for a daunting process. In an intensely competitive job market, university search committees seek candidates who can demonstrate vast skill sets beyond their field-specific expertise, from teaching to grant writing to lab management.
But Lugagne had a secret weapon to help him through the job search: his faculty mentor, Mary Dunlop. A Boston University College of Engineering associate professor of biomedical engineering, Dunlop guided Lugagne in refining his research proposal, gave him feedback on his interview materials, and even helped him organize a practice “chalk talk” with other faculty to ensure he was prepared for his presentations to search committees. The preparation paid off: Lugagne is joining the University of Oxford as an assistant professor of engineering science next year.
Now, in recognition of her deep commitment to the professional and personal development of postdoctoral researchers at BU, Dunlop has been awarded the University’s first-ever Award for Excellence in Mentoring Postdocs. She was honored at BU’s National Postdoc Appreciation celebration.
“These awards reflect the critical contributions of our postdocs and their mentors,” says Pallavi Eswara, director of postdoctoral affairs at BU. “Our postdocs are doing excellent research, but they are also contributing to our research community in countless other ways, as teachers, mentors, DEI leaders, and innovators.”
Read more here.
Congratulations Dr. Mary Dunlop!
Megan Hopton Receives the Synthetic Biology & Biotechnology Fellowship

Megan Hopton of the Hao Lab, recently received the Synthetic Biology & Biotechnology (SB2) Fellowship.
SB2's mission is to foster the next generation of researchers capable of rigorously and creatively applying synthetic biology principles and approaches to a broad range of scientific problems. In addition to building a field-defining synthetic biology curriculum that includes technical training, mastery of computational and data analysis methods, and critical thinking skills, SB2 provides robust training in the professional development skills required for success in this highly interdisciplinary field, as well as unique exposure into the biotechnology sector that is already being transformed by this field.
In Megan's research, she is developing nanosensors that respond to the activity of endogenous proteases by emitting bioorthogonal "synthetic biomarkers". She aims to leverage this highly multiplexable technology to create low-cost, noninvasive, point-of-care diagnostics that can be used to detect cancer or differentiate between disease etiologies. She is also interested in establishing a high-throughput approach to assess protease activity in a spatial context in order to better inform the development of protease-activatable diagnostics and therapeutics.
Congratulations Megan!
Congratulations to Soyoung Bae on American Crystallography Association Early Career Scientist Spotlight
We are thrilled to announce that Soyoung Bae, a PhD graduate student in the Molecular Biology, Cell Biology, and Biochemistry (MCBB) program, has been featured in the American Crystallographic Association's (ACA) Early Career Scientist Spotlight for 2024. This Spotlight not only provides insight into Soyoung’s background and experience, but extolls the facilities and instruments at Boston University’s Chemistry Department used for her work. This honor stemmed from her participation in the ACA Meeting in Baltimore MD in the summer of 2023, in which she was awarded the Journal of Chemical Crystallography poster prize.
Since joining Professor Tolan’s Laboratory in 2021 in collaboration with the Karen Allen lab in Chemistry, Soyoung has made remarkable strides in structural biology, focusing on enzyme mechanisms and ligand interactions through advanced techniques such as X-ray crystallography, surface plasmon resonance, and ultra-fast X-ray/spectroscopy. Her innovative work on protein conformational changes, as well as her effort solving nearly a hundred enzyme structures in complex with various small molecules has been instrumental in informing structure/activity relationships for a drug development project in collaboration with a small pharmaceutical company. All of which underscores her significant contributions to the field. Soyoung’s success has been supported by exceptional resources and guidance, notably from Dr. Jeff Bacon, who has provided invaluable expertise with the advanced instrumentation at the BU-Chemistry Instrumentation Center (CIC).
Please join us in celebrating Soyoung's achievements and in wishing her the best as she advances her career in structural biology.
Congratulations to Soyoung Bae on winning a Stanford PULSE Scholarship
Soyoung Bae, a third-year graduate student in the Molecular Biology, Cell Biology, and Biochemistry (MCBB) PhD Degree program, has been awarded the prestigious Stanford PULSE Scholarship for the Ultra-Fast X-ray Summer School (UXSS) in June 2024. The UXSS program, hosted by Stanford's PULSE Institute at the Stanford Linear Accelerator Center (SLAC) National Accelerator Laboratory, is renowned for its cutting-edge training in ultrafast (atto–femto second) X-ray science, and Soyoung’s selection is a testament to her exceptional research and potential in the field of structural biology. The award covered her lodging at the Stanford workshop, allowing her to participate in this highly esteemed program.
Soyoung’s research in the laboratory of Biology Professor Dean Tolan, where she employs sophisticated techniques such as X-ray crystallography and surface plasmon resonance, aligns perfectly with the innovative focus of UXSS. Her participation in this program will undoubtedly enhance her skills and knowledge, further advancing her impactful research on enzyme structure and function.
Beyond BU for the Summer: Alexandra Lion Completes Intensive Embryology Course

Alexandra Lion is a PhD candidate in the Bradham Lab, which studies pattern formation during embryonic development using the larval skeleton of the sea urchin Lytechinus variegatus as a model. Her projects focus on the role of the DEAD-box RNA helicase DDX6 in temporal regulation of development, and on the effect of per- and polyfluoryl alkyl substances (PFAS) on embryonic development and patterning.
Alexandra recently spent part of her summer attending the Marine Biological Laboratory course, Embryology: Concepts and Techniques in Modern Developmental Biology. The Embryology Course is an intensive six-week laboratory and lecture course for advanced graduate students, postdoctoral fellows, and more senior researchers who seek a broad and balanced view of modern issues in developmental biology.
Alexandra describes her experience below:
I applied to the Embryology course at the Marine Biological Laboratory because I wanted to expand my knowledge and connections within the field of developmental biology, as well as get more hands-on experience with a greater number of model organisms. Through this 6-week course, I was exposed to a wide range of well-established models such as Drosophila, zebrafish and C.elegans, as well as emerging model systems like acoels, cephalopods and ctenophores. I was able to greatly expand my microscopy experience since we were provided dozens of microscopes from vendors including Nikon, Leica, Andor and Zeiss. Having access to such a wide range of tools and organisms allowed me to try experiments that were outside of my prior expertise, such as lineage tracing, photo conversion, laser ablation, tissue grafting and more. This course really reinvigorated my sense of curiosity and opened my eyes to just how many unanswered questions there still are to probe. It helped that the course directors and faculty were enthusiastic about our ideas, and encouraged us to try any experiment we wanted to no matter how out-of-the-box. The faculty and course assistants frequently stayed in the lab with us students late into the night to help troubleshoot our experiments, and really supported all of our efforts.
The course introduced me to many people in the field, from other graduate students and postdocs to faculty who have been doing research for decades. Getting to work with the faculty in close quarters for up to a week at a time was incredibly valuable; you really get a feel for how the faculty are to work with, and how they might be as mentors. I also made a lot of really incredible friends through my time at Embryology; spending 6 weeks in the lab together, sharing all of our triumphs and setbacks at the bench, but also doing non-science things like practicing for the Embryology vs. Physiology softball game together really bonded us all. I’m grateful to have these people as both my friends and colleagues, and am excited to reunite at future conferences and events.
My attendance at the course was funded in part by a Max M. Burger Endowed Scholarship in Embryology and by the Society for Developmental Biology. Thank you also to the Biology Department and MCBB Program for their support, and to my advisor Cynthia Bradham for her encouragement and support of my attendance.